|
|
Contents
- Introduction
- Inputs & Outputs
- Exercises with web browser
- External links
- Exercises with curl and Perl
- Troubleshooting
Introduction
Proteins evolve by neutral mutations and natural selection. Therefore the network of sequence similarities is a rich source for mining homologous relationships. Most proteins are annotated using nearest neighbor inference.There are many many servers available to browse the network of homology relationships (for example at NCBI, EBI, Uniprot, and HHMI) but one has to wait up to a minute for results. SANSparallel was designed to suit those with a shorter span of attention.
This tutorial explains how to use the web interface to SANSparallel. SANSparallel takes a protein sequence as input and returns a set of similar sequences from Uniprot databases. The result is returned immediately. The retrieved sequences can be downloaded in FASTA format, aligned, and visualized in Jalview or Mview or as Sequence Logos (Figure 1). The web server can be accessed from:
- Home page: http://ekhidna2.biocenter.helsinki.fi/sans/index.html -> select Server tab
- Server URL: http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi

|
Inputs & Outputs
Sequence input is required. Public sequence databases can be accessed, for example, from NCBI or EBI and Uniprot. The query form offers two ways to enter the query sequence(s): type or copy-paste the sequences into the text box or upload a text file in FASTA format. This is an example of a sequence entry in FASTA format:>MCINX|MCINXTMP_001238-PA predicted protein MLFHTDDLQPIFETENRPDNFSERVSETQSDTPKGGETQPEIVTVGKEKKKGGKTFPCA QCGREFAHKNSLAYHTLMHGDKQQACRDDYRPCKCDECGRQFRQWSDLKYHKASLHSDK KQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKAFRASSYRLIHMRTHTGSKPY KCPQCDKGFRVSYDLQRHMHIHEKVRVKADDQKKTKDTKEKKQTITKTNEEKKEPESPS SEQIKSENRLPMLKSLLDKKPAKQSKKSPKKAPNVTVQNKIDEQFDEEIFDTRQDPYKF KEVYTNEKEFSNISHKFDRTDERELENLRSIKIPQIGETEDRNYSRENTDGKMQVFTQI DKGKEYSGPIVTNVVSLSDIRNLEREVLREPRVEIQGDGLENGFFERLSAFYNISAIThe query form is illustrated below.
|
Enter a protein sequence in FASTA format (example): or upload a FASTA file:
Number of hits (H):
1
10
50
100
250
500
1000
|
Example
- Click on this link for an active form, the figure above does not do anything.
- Copy-paste the example sequence from above into the active form.
- Set the value of H to 10.
- Set the search database to Swissprot.
- Click submit.
this link to repeat the search, the buttons in the figure above have been deactivated.
The result page has these four elements:
- The header line of the query protein is echoed in the result
- An array of post-processing tools can be launched from the table with light-blue background. Hovering the cursor above the question-mark icons gives usage tips. We will go through each tool in the exercises.
- The hits are listed in a table. Search metrics are reported on the left side and protein annotations on the right side. E-values above 1e-5 are flagged in orange and E-values above 0.1 are flagged in red. Very fast search mode does not compute alignments, so it shows only the Vote column on the left side. Uniprot/Swissprot annotations include Protein identifier, description, species and gene name. UniRef databases are annotated with cluster identifier, description, cluster size, and taxonomic range. Rows are sorted by bitscore (by vote in case of very fast search).
- The wall-clock time taken to process the request is reported last.
Exercises with web browser
- The result page links to two versions of Jalview. The following two steps are needed for setup.
- First, define preferences in Jalview Desktop. Click here to launch Jalview Desktop. Go to Tools -> Preferences -> Visual and uncheck Open file box. Go to Tools -> Preferences -> Colours, select Clustal as Alignment Colour and click OK. Preference settings are permanent. Once set, you won't need to change them again when you launch a new Jalview Desktop session.
- Check your Java security settings for Jalview Applet to run. On Windows 7: go to Control Panel -> Java (32-bit) -> General -> See the Security tab -> Enable Java content in the browser should be checked, security level should be High (minimum recommended) and http://ekhidna2.biocenter.helsinki.fi added to the Exception site list. With these settings, you should be able to pass Java security warnings by clicking Accept or OK or Run.
The setup steps above need only be done once and should be remembered by your computer thereafter. Now let's run through each of the post-processing buttons. We use the same example as above. You can click on this link to repeat the search.
- Click on the button labelled BLAST-like or try this direct link. You should get a result similar to the one below. The output is generated by running Pearson's fasta36 program against the hits reported by SANSparallel. The bitscores and e-values do not match exactly to those reported by SANSparallel; reasons are differences in gap scoring, database size and Karlin-Altschul parameters.
FASTA 36.3.6 Aug., 2014(preload9) Reference: W.R. Pearson & D.J. Lipman PNAS (1988) 85:2444-2448 Database: /tmp/49239.fa 10 sequences; 6132 total letters Query= Query Length=411 Score E Sequences producing significant alignments: (Bits) Value sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN= 132.2 8e-27sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN= 135.2 1e-27sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN= 97.1 3e-16sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN= 58.2 0.0002 sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Z 130.9 2e-26sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Z 127.9 2e-25sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Z 108.8 9e-20sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Z 73.8 3e-09sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Z 59.9 5e-05 sp|Q9Y6Q3|ZFP37_HUMAN Zinc finger protein 37 homolog OS=Homo sapi 127.0 3e-25sp|Q9Y6Q3|ZFP37_HUMAN Zinc finger protein 37 homolog OS=Homo sapi 95.8 7e-16sp|Q9Y6Q3|ZFP37_HUMAN Zinc finger protein 37 homolog OS=Homo sapi 70.3 3e-08 sp|Q9EPU7|Z354C_RAT Zinc finger protein 354C OS=Rattus norvegicus 126.6 4e-25sp|Q9EPU7|Z354C_RAT Zinc finger protein 354C OS=Rattus norvegicus 116.2 5e-22 sp|A6NNF4|ZN726_HUMAN Zinc finger protein 726 OS=Homo sapiens GN= 126.1 6e-25sp|A6NNF4|ZN726_HUMAN Zinc finger protein 726 OS=Homo sapiens GN= 140.0 4e-29sp|A6NNF4|ZN726_HUMAN Zinc finger protein 726 OS=Homo sapiens GN= 102.8 7e-18sp|A6NNF4|ZN726_HUMAN Zinc finger protein 726 OS=Homo sapiens GN= 62.5 9e-06sp|A6NNF4|ZN726_HUMAN Zinc finger protein 726 OS=Homo sapiens GN= 60.8 3e-05 sp|Q08AN1|ZN616_HUMAN Zinc finger protein 616 OS=Homo sapiens GN= 124.8 2e-24sp|Q08AN1|ZN616_HUMAN Zinc finger protein 616 OS=Homo sapiens GN= 139.1 8e-29sp|Q08AN1|ZN616_HUMAN Zinc finger protein 616 OS=Homo sapiens GN= 123.1 6e-24sp|Q08AN1|ZN616_HUMAN Zinc finger protein 616 OS=Homo sapiens GN= 117.9 2e-22 sp|Q8N823|ZN611_HUMAN Zinc finger protein 611 OS=Homo sapiens GN= 122.2 9e-24sp|Q8N823|ZN611_HUMAN Zinc finger protein 611 OS=Homo sapiens GN= 125.3 1e-24sp|Q8N823|ZN611_HUMAN Zinc finger protein 611 OS=Homo sapiens GN= 115.7 8e-22 sp|Q8N9K5|ZN565_HUMAN Zinc finger protein 565 OS=Homo sapiens GN= 119.7 4e-23sp|Q8N9K5|ZN565_HUMAN Zinc finger protein 565 OS=Homo sapiens GN= 99.7 4e-17sp|Q8N9K5|ZN565_HUMAN Zinc finger protein 565 OS=Homo sapiens GN= 65.6 8e-07 sp|Q8N988|ZN557_HUMAN Zinc finger protein 557 OS=Homo sapiens GN= 111.9 7e-21sp|Q8N988|ZN557_HUMAN Zinc finger protein 557 OS=Homo sapiens GN= 111.9 7e-21 sp|Q3SY52|ZIK1_HUMAN Zinc finger protein interacting with ribonuc 105.4 7e-19sp|Q3SY52|ZIK1_HUMAN Zinc finger protein interacting with ribonuc 104.5 1e-18 >sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN=ZNF728 PE=3 SV=1 Length=622 Score = 132.2 bits (297), Expect = 8e-27 Identities = 66/151 (43%), Positives = 92/151 (60%), Gaps = 9/151 (5%) Query 40 PEIVTVGKEKKKGGKTFPCAQCGREFAHKNSLAYHTLMHGDKQQACRDDYRPCKCDECGRQFRQW 104 P +T K G K + C +CG+ F ++L H ++H + +P KC+ECG+ F + Sbjct 380 PSSLTEHKRIHAGDKPYKCEECGKTFKWSSTLTKHKIIHTGE--------KPYKCEECGKAFTTF 436 Query 105 SDLKYHKASLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKAFRASSYRLIHMR 169 S L HK +H+ +K +KC+ CGK F+ SL+ H+ IH GEK YKCE C KAF+ SS + H R Sbjct 437 SSLTKHKV-IHTGEKHYKCEECGKVFSWSSSLTTHKAIHAGEKLYKCEECGKAFKWSSNLMEHKR 500 Query 170 THTGSKPYKCPQCDKGFRVSYDLQRHMHIH 199 HTG KPYKC +C K F +L +H IH Sbjct 501 IHTGEKPYKCEECGKAFSKVANLTKHKVIH 530 // snip // Score = 104.5 bits (233), Expect = 1e-18 Identities = 44/106 (41%), Positives = 62/106 (58%), Gaps = 1/106 (0%) Query 93 KCDECGRQFRQWSDLKYHKASLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKA 157 K ECG+ R YH +++ KK ++C CGK F +YSL H+R+HTGE+ ++C C K Sbjct 212 KSGECGKASRHKHTPVYHPR-VYTGKKLYECSKCGKAFRGKYSLVQHQRVHTGERPWECNECGKF 275 Query 158 FRASSYRLIHMRTHTGSKPYKCPQCDKGFRVSYDLQRHMHIH 199 F +S+ H R HTG +PY+C +C K FR + L H IH Sbjct 276 FSQTSHLNDHRRIHTGERPYECSECGKLFRQNSSLVDHQKIH 317 Lambda K H 0.298 0.131 0.428 Gapped Lambda 0.298 0.131 0.428 Effective search space used: 10 Database: /tmp/49239.fa Number of letters in database: 6132 Number of sequences in database: 10 Matrix: BL62 Gap Penalties: Existence: -10, Extension: -2Elapsed time: 0.432391881942749 seconds - Click on the button labelled FASTA in the Profile row or try this direct link. Profile means a stacked alignment, where the hits are stacked onto the query sequence and insertions are thrown away. Deletions and unaligned ends are padded with gap characters ('-'). You should get a result similar to the one below. This is a gapped alignment in FASTA format and it could be copy-pasted to many alignment editing-visualization tools. Our web server has a direct link to Jalview (Figure 1).
>Query MLFHTDDLQPIFETENRPDNFSERVSETQSDTPKGGETQPEIVTVGKEKKKGGKTFPCAQCGREFAHKNSLAYHTLMHGDKQQACRDDYRPCKCDECGRQFRQWSDLKYHKASLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKAFRASSYRLIHMRTHTGSKPYKCPQCDKGFRVSYDLQRHMHIHEKVRVKADDQKKTKDTKEKKQTITKTNEEKKEPESPSSEQIKSENRLPMLKSLLDKKPAKQSKKSPKKAPNVTVQNKIDEQFDEEIFDTRQDPYKFKEVYTNEKEFSNISHKFDRTDERELENLRSIKIPQIGETEDRNYSRENTDGKMQVFTQIDKGKEYSGPIVTNVVSLSDIRNLEREVLREPRVEIQGDGLENGFFERLSAFYNISAI >sp|Q6ZR52|ZN493_HUMAN Zinc finger protein 493 OS=Homo sapiens GN=ZNF493 PE=2 SV=3 -----------------------------------------------------KPYKCEECGKAFKRSSTLTKHRIIHTEE--------KPYKCEECGKAFNQSSTLSIHKI-IHTGEKPYKCEECGKAFKRSSTLTIHKMIHTGEKPYKCEECGKAFNRSSHLTTHKRIHTGHKPYKCKECGKSFSVFSTLTKHKIIHDKKPYKCEECGKAFNRSSILSIHKKIHTEKPYKCEECGKAFKRSSHLAGHKQIHSVQKPYKCEECGKAFSIFSTLTK------HKIIHTEEKPYKCEKCGKTFYRFSNLNHKIIHTGEK--------------------------------------------------------------------------------------------- >sp|A6NN14|ZN729_HUMAN Zinc finger protein 729 OS=Homo sapiens GN=ZNF729 PE=2 SV=4 ---------------------------------------------------GEKPYKCEECGKAFKWSSKLTVHKVVHTGE--------KPYKCEECGKAFSQFSTLKKHKI-IHTGKKPYKCEECGKAFNSSSTLMKHKIIHTGEKPYKCEECGKAFRQSSHLTRHKAIHTGEKPYKCEECGKAFNHFSDLRRHKIIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- >sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN=ZNF728 PE=3 SV=1 ---------------------------------------PSSLTEHKRIHAGDKPYKCEECGKTFKWSSTLTKHKIIHTGE--------KPYKCEECGKAFTTFSSLTKHKV-IHTGEKHYKCEECGKVFSWSSSLTTHKAIHAGEKLYKCEECGKAFKWSSNLMEHKRIHTGEKPYKCEECGKAFSKVANLTKHKVIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- >sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Zfp11 PE=2 SV=2 -----------------------------------------VLTQHRITHTGEKPFKCKECGRAFKYNSTLTQHEVIHTEA--------KPYRCQECGKAFKRSHTLSQHQV-IHKGEKPHKCDECGRAFSKHSSLTQHQVIHTGEKPYQCRECGKAFRYQSTLTRHHIVHTGAKPYKCPECDKAFNNSSTLSRHQIIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- >sp|Q8IYI8|ZN440_HUMAN Zinc finger protein 440 OS=Homo sapiens GN=ZNF440 PE=2 SV=1 -------------------------------------TCPRYVRIHERTHSRKNLYECKQCGKALSSLTSFQTHVRLHSGE--------RPYECKICGKDFCSVNSFQRHE-KIHSGEKPYKCKQCGKAFPHSSSLRYHERTHTGEKPYECKQCGKAFRSASHLRVHGRTHTGEKPYECKECGKAFRYVNNLQSHERTQTHIRIHSGERRKCKICGKGFYCPKSFQRHEKTHTGEKLYECKQRSVVPSVVPVFDIMKGLTLERSPINASNVGKPSELCQSFECMVGLTKRNPMSV----SNDGKPSDLPHTFEHTMERSPMHVRNVGNP---------------------------------------------------------------------------------- >sp|P17026|ZNF22_HUMAN Zinc finger protein 22 OS=Homo sapiens GN=ZNF22 PE=1 SV=3 -----------------------------------------------------KPYKCTECEKSFSQSSTLFQHQKIHTGKKSH--------KCADCGKSFFQSSNLIQHRR-IHTGEKPYKCDECGESFKQSSNLIQHQRIHTGEKPYQCDECGRCFSQSSHLIQHQRTHTGEKPYQCSECGKCFSQSSHLRQHMKVHKE------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ >sp|Q6NX49|ZN544_HUMAN Zinc finger protein 544 OS=Homo sapiens GN=ZNF544 PE=2 SV=1 ----------------------------------------ELVT-HKRTHTGEKPFKCTQCGKSFSQKYDLVVHQRTHTGE--------KPYECNLCGKSFSQSSKLITHQR-IHTGEKPYQCIECGKSFRWNSNLVIHQRIHTGEKPYDCTHCGKSFSQSYQLVAHKRTHTGEKPYECNECGKAFNRSTQLIRHLQIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- >sp|Q571J5|Z354C_MOUSE Zinc finger protein 354C OS=Mus musculus GN=Znf354c PE=2 SV=2 -----------------------------------GFTQSLHLLEHKRLHTGEKPYKCSECGKSFSHRSSLLAHQRTHTGE--------KPYKCSECEKAFGSSSTLIKH-LRVHTGEKPYRCRQCGKAFSQCSTLTVHQRIHTGEKLYKCAECDKAFNCRAKLHRHQRIHTGEKPYKCAECGKGYSQFPSLAEHQRLH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- >sp|Q8NB42|ZN527_HUMAN Zinc finger protein 527 OS=Homo sapiens GN=ZNF527 PE=2 SV=2 ---------------------------------------------------GEKPFACNECGKAFSRYAFLVEHQRIHTGE--------KPYECKECNKAFRQSAHLNQHQR-IHTGEKPYECNQCGKAFSRRIALTLHQRIHTGEKPFKCSECGKTFGYRSHLNQHQRIHTGEKPYECIKCGKFFRTDSQLNRHHRIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- >sp|Q8NEM1|ZN680_HUMAN Zinc finger protein 680 OS=Homo sapiens GN=ZNF680 PE=2 SV=2 -----------------------------------------------------KPFKCEECGKAFSLFSILSKHKIIHGDKPYKCDECHKPFKCEECGKDFNQFSNLTKHK-KIHTGEKPYKCEECGKAFNQFANLTRHKKIHTGEKSYKCEECGKAFIQSSNLTEHMRIHTGEKPYKCEECGKAFNGCSSLTRHKRIHTR------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
- Click on the button labelled Desktop in the Profile row or try this direct link. This launches a Jalview Desktop session with the previous gapped FASTA alignment automatically loaded (Figure 3).
Figure 3. A screenshot from Jalview Desktop.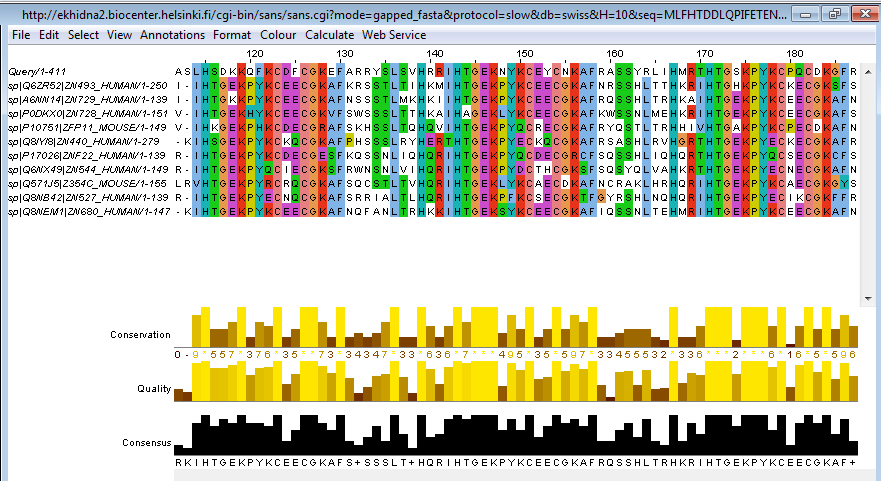
- Click on the button labelled Applet in the Profile row or try this direct link. This launches a Jalview Applet session with the previous gapped FASTA alignment automatically loaded, when you click the button labelled 'Start Jalview'. The functionality of the applet is less extensive than that of Jalview Desktop. The alignment part looks the same as in Jalview Desktop (Figure 3), but the menus have fewer options.
- Click on the button labelled Mview in the Profile row or try this direct link. You should get a similar result to the one shown below. Mview output is pure HTML and provides an alternative to Java based Jalview though Mview is rather slow on large alignments.
0Reference sequence (1): Query Identities normalised by aligned length. Colored by: consensus/70% and property
1 Query 100.0% MLFHTDDLQPIFETENRPDNFSERVSETQSDTPKGGETQPEIVTVGKEKKKGGKTFPCAQCGREFAHKNSLAYHTLMHGDKQQACRDDYRPCKCDECGRQFRQWSDLKYHKASLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKAFRASSYRLIHMRTHTGSKPYKCPQCDKGFRVSYDLQRHMHIHEKVRVKADDQKKTKDTKEKKQTITKTNEEKKEPESPSSEQIKSENRLPMLKSLLDKKPAKQSKKSPKKAPNVTVQNKIDEQFDEEIFDTRQDPYKFKEVYTNEKEFSNISHKFDRTDERELENLRSIKIPQIGETEDRNYSRENTDGKMQVFTQIDKGKEYSGPIVTNVVSLSDIRNLEREVLREPRVEIQGDGLENGFFERLSAFYNISAI 2 sp|Q6ZR52|ZN493_HUMAN Zinc finger protein 493 OS=... 33.2% -----------------------------------------------------KPYKCEECGKAFKRSSTLTKHRIIHTEE--------KPYKCEECGKAFNQSSTLSIHKI-IHTGEKPYKCEECGKAFKRSSTLTIHKMIHTGEKPYKCEECGKAFNRSSHLTTHKRIHTGHKPYKCKECGKSFSVFSTLTKHKIIHDKKPYKCEECGKAFNRSSILSIHKKIHTEKPYKCEECGKAFKRSSHLAGHKQIHSVQKPYKCEECGKAFSIFSTLTK------HKIIHTEEKPYKCEKCGKTFYRFSNLNHKIIHTGEK--------------------------------------------------------------------------------------------- 3 sp|A6NN14|ZN729_HUMAN Zinc finger protein 729 OS=... 45.9% ---------------------------------------------------GEKPYKCEECGKAFKWSSKLTVHKVVHTGE--------KPYKCEECGKAFSQFSTLKKHKI-IHTGKKPYKCEECGKAFNSSSTLMKHKIIHTGEKPYKCEECGKAFRQSSHLTRHKAIHTGEKPYKCEECGKAFNHFSDLRRHKIIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- 4 sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=... 41.2% ---------------------------------------PSSLTEHKRIHAGDKPYKCEECGKTFKWSSTLTKHKIIHTGE--------KPYKCEECGKAFTTFSSLTKHKV-IHTGEKHYKCEECGKVFSWSSSLTTHKAIHAGEKLYKCEECGKAFKWSSNLMEHKRIHTGEKPYKCEECGKAFSKVANLTKHKVIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- 5 sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=M... 41.1% -----------------------------------------VLTQHRITHTGEKPFKCKECGRAFKYNSTLTQHEVIHTEA--------KPYRCQECGKAFKRSHTLSQHQV-IHKGEKPHKCDECGRAFSKHSSLTQHQVIHTGEKPYQCRECGKAFRYQSTLTRHHIVHTGAKPYKCPECDKAFNNSSTLSRHQIIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- 6 sp|Q8IYI8|ZN440_HUMAN Zinc finger protein 440 OS=... 28.8% -------------------------------------TCPRYVRIHERTHSRKNLYECKQCGKALSSLTSFQTHVRLHSGE--------RPYECKICGKDFCSVNSFQRHE-KIHSGEKPYKCKQCGKAFPHSSSLRYHERTHTGEKPYECKQCGKAFRSASHLRVHGRTHTGEKPYECKECGKAFRYVNNLQSHERTQTHIRIHSGERRKCKICGKGFYCPKSFQRHEKTHTGEKLYECKQRSVVPSVVPVFDIMKGLTLERSPINASNVGKPSELCQSFECMVGLTKRNPMSV----SNDGKPSDLPHTFEHTMERSPMHVRNVGNP---------------------------------------------------------------------------------- 7 sp|P17026|ZNF22_HUMAN Zinc finger protein 22 OS=H... 39.2% -----------------------------------------------------KPYKCTECEKSFSQSSTLFQHQKIHTGKKSH--------KCADCGKSFFQSSNLIQHRR-IHTGEKPYKCDECGESFKQSSNLIQHQRIHTGEKPYQCDECGRCFSQSSHLIQHQRTHTGEKPYQCSECGKCFSQSSHLRQHMKVHKE------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ 8 sp|Q6NX49|ZN544_HUMAN Zinc finger protein 544 OS=... 41.5% ----------------------------------------ELVT-HKRTHTGEKPFKCTQCGKSFSQKYDLVVHQRTHTGE--------KPYECNLCGKSFSQSSKLITHQR-IHTGEKPYQCIECGKSFRWNSNLVIHQRIHTGEKPYDCTHCGKSFSQSYQLVAHKRTHTGEKPYECNECGKAFNRSTQLIRHLQIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- 9 sp|Q571J5|Z354C_MOUSE Zinc finger protein 354C OS... 39.0% -----------------------------------GFTQSLHLLEHKRLHTGEKPYKCSECGKSFSHRSSLLAHQRTHTGE--------KPYKCSECEKAFGSSSTLIKH-LRVHTGEKPYRCRQCGKAFSQCSTLTVHQRIHTGEKLYKCAECDKAFNCRAKLHRHQRIHTGEKPYKCAECGKGYSQFPSLAEHQRLH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- 10 sp|Q8NB42|ZN527_HUMAN Zinc finger protein 527 OS=... 41.2% ---------------------------------------------------GEKPFACNECGKAFSRYAFLVEHQRIHTGE--------KPYECKECNKAFRQSAHLNQHQR-IHTGEKPYECNQCGKAFSRRIALTLHQRIHTGEKPFKCSECGKTFGYRSHLNQHQRIHTGEKPYECIKCGKFFRTDSQLNRHHRIH-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- 11 sp|Q8NEM1|ZN680_HUMAN Zinc finger protein 680 OS=... 47.3% -----------------------------------------------------KPFKCEECGKAFSLFSILSKHKIIHGDKPYKCDECHKPFKCEECGKDFNQFSNLTKHK-KIHTGEKPYKCEECGKAFNQFANLTRHKKIHTGEKSYKCEECGKAFIQSSNLTEHMRIHTGEKPYKCEECGKAFNGCSSLTRHKRIHTR------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
Elapsed time: 1.21434283256531 seconds
- Click on the button labelled Logo in the Aligned row or try this direct link. You will be redicted to the Skylign server in a moment. You should see a dynamic sequence logo similar to the static picture below.
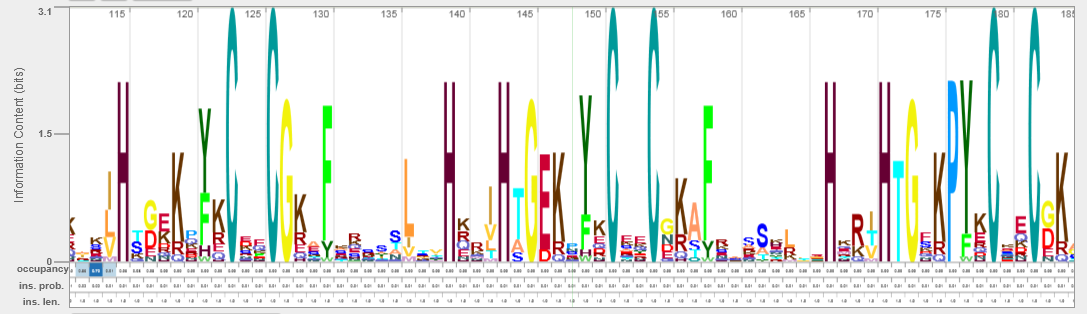
- Click on the button labelled FASTA in the Unaligned row or try this direct link. You should get a result similar to the one below. This FASTA file contains the full amino acid sequences of the query and all the accepted hits. The FASTA file could be copy-pasted to many multiple sequence alignment tools. Our web server has a direct link to Jalview Desktop (Figure 1).
>MCINX|MCINXTMP_001238-PA predicted protein MLFHTDDLQPIFETENRPDNFSERVSETQSDTPKGGETQPEIVTVGKEKKKGGKTFPCAQCGREFAHKNSLAYHTLMHGDKQQACRDDYRPCKCDECGRQFRQWSDLKYHKASLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKAFRASSYRLIHMRTHTGSKPYKCPQCDKGFRVSYDLQRHMHIHEKVRVKADDQKKTKDTKEKKQTITKTNEEKKEPESPSSEQIKSENRLPMLKSLLDKKPAKQSKKSPKKAPNVTVQNKIDEQFDEEIFDTRQDPYKFKEVYTNEKEFSNISHKFDRTDERELENLRSIKIPQIGETEDRNYSRENTDGKMQVFTQIDKGKEYSGPIVTNVVSLSDIRNLEREVLREPRVEIQGDGLENGFFERLSAFYNISAI >sp|Q8N9K5|ZN565_HUMAN Zinc finger protein 565 OS=Homo sapiens GN=ZNF565 PE=2 SV=2 MRRGPWERWSLASHRLDAGLCTCPREESREIRAGQIVLKAMAQGLVTFRDVAIEFSLEEWKCLEPAQRDLYREVTLENFGHLASLGLSISKPDVVSLLEQGKEPWMIANDVTGPWCPDLESRCEKFLQKDIFEIGAFNWEIMESLKCSDLEGSDFRADWECEGQFERQVNEECYFKQVNVTYGHMPVFQHHTSHTVRQSRETGEKLMECHECGKAFSRGSHLIQHQKIHTGEKPFGCKECGKAFSRASHLVQHQRIHTGEKPYDCKDCGKAFGRTSELILHQRLHTGVKPYECKECGKTFRQHSQLILHQRTHTGEKPYVCKDCGKAFIRGSQLTVHRRIHTGARPYECKECGKAFRQHSQLTVHQRIHTGEKPYECKECGKGFIHSSEVTRHQRIHSGEKPYECKECGKAFRQHAQLTRHQRVHTGDRPYECKDCGKAFSRSSYLIQHQRIHTGDKPYECKECGKAFIRVSQLTHHQRIHTCEKPYECRECGMAFIRSSQLTEHQRIHPGIKPYECRECGQAFILGSQLIEHYRIHTG >sp|A6NNF4|ZN726_HUMAN Zinc finger protein 726 OS=Homo sapiens GN=ZNF726 PE=2 SV=4 MGLLTFRDVAIEFSLEEWQCLDTAQKNLYRNVMLENYRNLAFLGIAVSKPDLIICLEKEKEPWNMKRDEMVDEPPGICPHFAQDIWPEQGVEDSFQKVILRRFEKCGHENLQLRKGCKSVDECKVHKEGYNGLNQCFTTTQGKASQCGKYLKVFYKFINLNRYKIRHTRKKPFKCKNCVKSFCMFSHKTQHKSIYTTEKSYKCKECGKTFNWSSTLTNHKKTHTEEKPYKCEEYGKAFNQSSNYTTHKVTHTGEKPYKCEECGKAFSQSSTLTIHKRIHTGEKPCKCEECGKAFSQPSALTIHKRMHIGEKPYKCEECGKAFVWSSTLTRHKRLHSGEKPYKCEECAKAFSQFGHLTTHRIIHTGEKPYKCEECGKAFIWPSTLTKHKRIHTGEKPYKCEECGKAFHRSSNLTKHKIIHTGEKPYKCEECGKAFIWSSNLTEHKKIHTREKPYKCEECSKAFSRSSALTTHKRMHTGEKPYKCEECGKAFSQSSTLTAHKIIHTGEKPYKCEECGKAFILSSTLSKHKRIHTGEKPYKCEECGKTFNQSSNLSTHKIIHTGEKPYKCEECGKAFNRSSNLSTHKIIHTGEKPYKCDECGKSFIWSSTLFKHKRIHTGEKPYKCEECGKAFNHSQILLHIRHKRMHTGEKPYKCEECGKSFNLSSTFIKHKVIHTGVKLYKCEECGKVFFWSSALTRHKKIHAGQQPYKWEKIGKAFNQSSHLTTDKITHIGEKSYKCE >sp|Q9Y6Q3|ZFP37_HUMAN Zinc finger protein 37 homolog OS=Homo sapiens GN=ZFP37 PE=2 SV=3 MSVSSGVQILTKPETVDRRRSAETTKEAGRPLEMAVSEPEASAAEWKQLDPAQSNLYNDVMLENYCNQASMGCQAPKPDMISKLEKGEAPWLGKGKRPSQGCPSKIARPKQKETDGKVQKDDDQLENIQKSQNKLLREVAVKKKTQAKKNGSDCGSLGKKNNLHKKHVPSKKRLLKFESCGKILKQNLDLPDHSRNCVKRKSDAAKEHKKSFNHSLSDTRKGKKQTGKKHEKLSSHSSSDKCNKTGKKHDKLCCHSSSHIKQDKIQTGEKHEKSPSLSSSTKHEKPQACVKPYECNQCGKVLSHKQGLIDHQRVHTGEKPYECNECGIAFSQKSHLVVHQRTHTGEKPYECIQCGKAHGHKHALTDHLRIHTGEKPYECAECGKTFRHSSNLIQHVRSHTGEKPYECKECGKSFRYNSSLTEHVRTHTGEIPYECNECGKAFKYSSSLTKHMRIHTGEKPFECNECGKAFSKKSHLIIHQRTHTKEKPYKCNECGKAFGHSSSLTYHMRTHTGESPFECNQCGKGFKQIEGLTQHQRVHTGEKPYECNECGKAFSQKSHLIVHQRTHTGEKPYECNECEKAFNAKSQLVIHQRSHTGEKPYECNECGKTFKQNASLTKHVKTHSEDKSHE >sp|Q8N988|ZN557_HUMAN Zinc finger protein 557 OS=Homo sapiens GN=ZNF557 PE=2 SV=2 MAAVVLPPTAASQREGHTEGGELVNELLKSWLKGLVTFEDVAVEFTQEEWALLDPAQRTLYRDVMLENCRNLASLGNQVDKPRLISQLEQEDKVMTEERGILSGTCPDVENPFKAKGLTPKLHVFRKEQSRNMKMERNHLGATLNECNQCFKVFSTKSSLTRHRKIHTGERPYGCSECGKSYSSRSYLAVHKRIHNGEKPYECNDCGKTFSSRSYLTVHKRIHNGEKPYECSDCGKTFSNSSYLRPHLRIHTGEKPYKCNQCFREFRTQSIFTRHKRVHTGEGHYVCNQCGKAFGTRSSLSSHYSIHTGEYPYECHDCGRTFRRRSNLTQHIRTHTGEKPYTCNECGKSFTNSFSLTIHRRIHNGEKSYECSDCGKSFNVLSSVKKHMRTHTGKKPYECNYCGKSFTSNSYLSVHTRMHNRQM >sp|Q3SY52|ZIK1_HUMAN Zinc finger protein interacting with ribonucleoprotein K OS=Homo sapiens GN=ZIK1 PE=2 SV=1 MAAAALRAPTQVTVSPETHMDLTKGCVTFEDIAIYFSQDEWGLLDEAQRLLYLEVMLENFALVASLGCGHGTEDEETPSDQNVSVGVSQSKAGSSTQKTQSCEMCVPVLKDILHLADLPGQKPYLVGECTNHHQHQKHHSAKKSLKRDMDRASYVKCCLFCMSLKPFRKWEVGKDLPAMLRLLRSLVFPGGKKPGTITECGEDIRSQKSHYKSGECGKASRHKHTPVYHPRVYTGKKLYECSKCGKAFRGKYSLVQHQRVHTGERPWECNECGKFFSQTSHLNDHRRIHTGERPYECSECGKLFRQNSSLVDHQKIHTGARPYECSQCGKSFSQKATLVKHQRVHTGERPYKCGECGNSFSQSAILNQHRRIHTGAKPYECGQCGKSFSQKATLIKHQRVHTGERPYKCGDCGKSFSQSSILIQHRRIHTGARPYECGQCGKSFSQKSGLIQHQVVHTGERPYECNKCGNSFSQCSSLIHHQKCHNT >sp|Q9EPU7|Z354C_RAT Zinc finger protein 354C OS=Rattus norvegicus GN=Znf354c PE=1 SV=1 MAVDLLAARGTEPVTFRDVAVSFSQDEWLHLDPAQRTLYREVMLENYSNLASLGFQASIPPVIGKLQKGQDPCMEREAPEDTCLDFQIQSEIEASSPEQDVFIEGPSRGLLKNRSTKCAYWKISFGELVKYERLETAQEQEKKAHEPGAASPKEVTSEDGIPTDPELEKPLFMNKALVSQETDPIERVPGMYHTSEKDLPQDFDLMRNFQIYPGQKPYVCSECGKGFSQSLHLLEHKRIHTGEKPYKCSECGKSFSHRSSLLAHQRTHTGEKPYKCSECEKAFGSSSTLIKHLRVHTGEKPYRCRECGKAFSQCSTLTVHQRIHTGEKLYKCAECDKAFNCRAKLHRHQRIHTGEKPYKCAECGKGYSQFPSLAEHQRLHTGGQLCQCLQCGRTFTRVSTLIEHQRIHTGQKPYQCNECGKTFNQYSSFNEHRKIHTGEKLYTCEECGKAFGCKSNLYRHQRIHTGEKPYQCNQCGKAFSQYSFLTEHERIHTGEKLYKCMECGKAYSYRSNLCRHKKVHLKERLYKWKEYGTPFMYGSSLAPHQRCLKGEKPEDLNSSL >sp|Q08AN1|ZN616_HUMAN Zinc finger protein 616 OS=Homo sapiens GN=ZNF616 PE=2 SV=2 MATQGHLTFKDVAIEFSQEEWKCLEPVQKALYKDVMLENYRNLVFLGISPKCVIKELPPTENSNTGERFQTVALERHQSYDIENLYFREIQKHLHDLEFQWKDGETNDKEVPVPHENNLTGKRDQHSQGDVENNHIENQLTSNFESRLAELQKVQTEGRLYECNETEKTGNNGCLVSPHIREKTYVCNECGKAFKASSSLINHQRIHTTEKPYKCNECGKAFHRASLLTVHKVVHTRGKSYQCDVCGKIFRKNSYFVRHQRSHTGQKPYICNECGKSFSKSSHLAVHQRIHTGEKPYKCNLCGKSFSQRVHLRLHQTVHTGERPFKCNECGKTFKRSSNLTVHQVIHAGKKPYKCDVCGKAFRHRSNLVCHRRIHSGEKQYKCNECGKVFSKRSSLAVHRRIHTVEKPCKCNECGKVFSKRSSLAVHQRIHTGQKTYKCNKCGKVYSKHSHLAVHWRIHTGEKAYKCNECGKVFSIHSRLAAHQRIHTGEKPYKCNECGKVFSQHSRLAVHRRIHTGEKPYKCKECGKVFSDRSAFARHRRIHTGEKPYKCKECGKVFSQCSRLTVHRRIHSGEKPYKCNECGKVYSQYSHLVGHRRVHTGEKPYKCHECGKAFNQGSTLNRHQRIHTGEKPYKCNQCGNSFSQRVHLRLHQTVHTGDRPYKCNECGKTFKRSSNLTAHQIIHAGKKPYKCDECGKVFRHSSHLVSHQRIHTGEKRYKCIECGKAFGRLFSLSKHQRIHSGKKPYKCNECGKSFICRSGLTKHRIRHTGESLTTKLNVTRP >sp|P0DKX0|ZN728_HUMAN Zinc finger protein 728 OS=Homo sapiens GN=ZNF728 PE=3 SV=1 MGSLTFRDVAIQFSLEEWQCLDTAQQNLYRNVMLENYRNLVFLGIAAPKPDLIIFLEQGKEPWNMKRHELVKEPPVICSHFAQDLWPEQGREDSFQKVILRRYEKCGHENLQLKIGCTNVDECKVHKKGYNKLNQSLTTTQSKVFQCGKYANIFHKCSNSKRHKIRHTGKKLLKCKEYVRSFCMLSHLSQHKRIYTRENSYKSEEHGKAFNWSSALTYKRIHTGEKPCKCEECGKAFSKFSILTKHKVIHTGEKHYKCEECGKAFTRSSSLIEHKRSHAGEKPYKCEECGKAFSKASTLTAHKTIHAGEKPYKCEECGKAFNRSSNLMEHKRIHTGEKPCKCEECGKAFGNFSTLTKHKVIHTGEKPYKCEECGKAFSWPSSLTEHKRIHAGDKPYKCEECGKTFKWSSTLTKHKIIHTGEKPYKCEECGKAFTTFSSLTKHKVIHTGEKHYKCEECGKVFSWSSSLTTHKAIHAGEKLYKCEECGKAFKWSSNLMEHKRIHTGEKPYKCEECGKAFSKVANLTKHKVIHTGEKQYKCEECGKAFIWSSRLSEHKRIHTGEKPYKCEECGKAFSWVSVLNKHKKIHAGKKFYKCEECGKDFNQSSHLTTHKRIHTGGKTLQM >sp|Q8N823|ZN611_HUMAN Zinc finger protein 611 OS=Homo sapiens GN=ZNF611 PE=2 SV=2 MLREEAAQKRKGKEPGMALPQGRLTFRDVAIEFSLAEWKCLNPSQRALYREVMLENYRNLEAVDISSKCMMKEVLSTGQGNTEVIHTGTLQRHESHHIGDFCFQEIEKEIHDIEFQCQEDERNGLEAPMTKIKKLTGSTDQHDHRHAGNKPIKDQLGSSFYSHLPELHIFQIKGEIGNQLEKSTNDAPSVSTFQRISCRPQTQISNNYGNNPLNSSLLPQKQEVHMREKSFQCNKSGKAFNCSSLLRKHQIPHLGDKQYKCDVCGKLFNHEQYLACHDRCHTVEKPYKCKECGKTFSQESSLTCHRRLHTGVKRYNCNECGKIFGQNSALLIDKAIDTGENPYKCNECDKAFNQQSQLSHHRIHTGEKPYKCEECDKVFSRKSTIETHKRIHTGEKPYRCKVCDTAFTWHSQLARHRRIHTAKKTYKCNECGKTFSHKSSLVCHHRLHGGEKSYKCKVCDKAFVWSSQLAKHTRIDCGEKPYKCNECGKTFGQNSDLLIHKSIHTGEQPYKCDECEKVFSRKSSLETHKIGHTGEKPYKCKVCDKAFACHSYLAKHTRIHSGEKPYKCNECSKTFSHRSYLVCHHRVHSGEKPYKCNECSKTFSRRSSLHCHRRLHSGEKPYKCNECGNTFRHCSSLIYHRRLHTGEKSYKCTICDKAFVRNSLLSRHTRIHTAEKPYKCNECGKAFNQQSHLSRHHRIHTGEKP >sp|P10751|ZFP11_MOUSE Zinc finger protein 11 OS=Mus musculus GN=Zfp11 PE=2 SV=2 MSPENLSDCNNSVKDFDQHPELTIRQCVHREKPYKQEECDDSACDQHLRVHKGGMPYECKDCGKAFKYRSVLYQHRIIHTAARPYKCKECGKAFKRSRNLAQHQVTHKREKPHKCEECGRAFSALSVLTQHRITHTGEKPFKCKECGRAFKYNSTLTQHEVIHTEAKPYRCQECGKAFKRSHTLSQHQVIHKGEKPHKCDECGRAFSKHSSLTQHQVIHTGEKPYQCRECGKAFRYQSTLTRHHIVHTGAKPYKCPECDKAFNNSSTLSRHQIIHTGEKPYKCQECGRAFYCSSFLIQHMKIHFEEMPYRCRECGKPFRLSSQLIRHQRIHTGEEPYICRECGKTFKYQSNLTRHQILHTGAKPYKCPECGKAFNNSSTLTRHQIIHTGEKPYKCQECSKAFYCSSYLIQHMKIHFEEIPYRCKECGKPFKCSSQLIRHQRIHSGEKPYICKECGKAFNCTSYLTKHQRIHTGEKPYVCQECGKAFNCSSYLSKHQRIHIGDRLYKCKECGKAYYFSSQLNRHQRIHTGEKPYVCQECGKAFNCSSYLTKHQRIHIVEKPYVCKECSKAFSCSSYLTKHQRIHAGDRLYKCTECGKAYCFSSQLNRHQRIHTRERPYRCKECGKAFITSSCLKRHQETHTLQTPNSV
- Click on the button labelled Desktop in the Profile row or try this direct link. This launches a Jalview Desktop session with the previous gapped FASTA alignment automatically loaded. Note that the sequences as loaded are unaligned (Figure 5a). You must generate an alignment using the Web Services -> Alignment options -> Run Muscle with preset -> Large alignments (balanced). Make sure to select or deselect all sequences. Figure 5b shows the result of multiple sequence aligment. Muscle is recommended because it is one of the fastest tools also on large alignments. The Alignment menu is inactive (shown by grey colour) while Jalview is loading JABA services. The active menu is written in black colour. Jalview Applet loads faster than Jalview Desktop but the applet does not support Alignment.
Figure 5a. Snapshot of unaligned sequences loaded into Jalview Desktop.
Figure 5b. Snapshot of the same sequences as in Figure 4 after multiple sequence alignment with Muscle.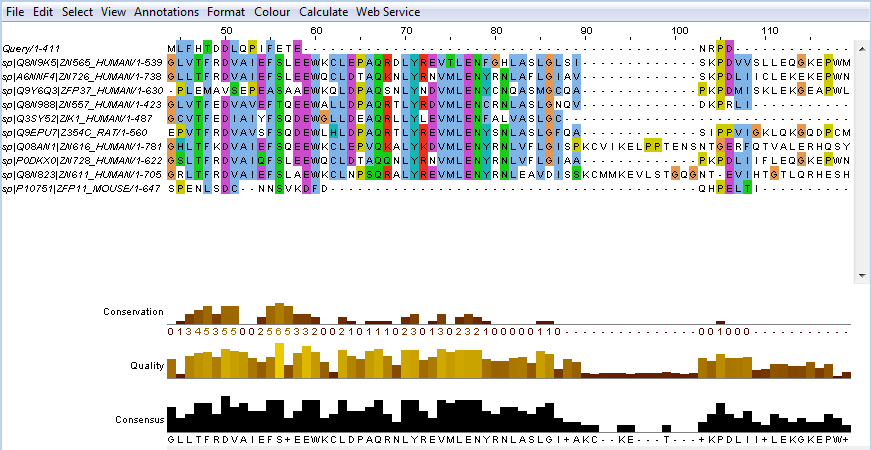
This concludes our tour through each of the post-processing buttons.
- Sequence similarity searches are often done for the purpose of function assignment and conservation patterns are examined to validate homology.
- The butterfly protein used as example has consistent zinc finger hits in Swissprot, but they are from somewhat distant species. Do a search against Uniprot and study the annotations of the hits. Now the closest hits are from other butterflies and have higher sequence similarity.
- There are uncharacterized proteins mixed among proteins annotated as zinc fingers. View the hits' profile in Jalview. Pick the invariably conserved residues from the Conservation panel. All hits match a tandemly repeated C2H2 signature. It is evident that all these sequences are zinc finger proteins.
- The profile view is often sufficient to identify conserved sequence patterns. An accurate alignment is a pre-requisite for phylogenetic analysis. Multiple sequence alignment can increase the accuracy of alignment, especially if the sequences are distantly related. Generate a multiple sequence alignment in Jalview Desktop using Muscle, MAFFT and Clustal. Are there differences?
- Compare the speed of SANSparallel to similar servers:
External links
The CGI script can be called to generate any of the output formats depicted in Figure 1. The parameters of the CGI script are summarized below.| Key | Description | Default | Values |
|---|---|---|---|
| db | name of database | uniprot |
|
| mode | type of output | table |
|
| H | number of hits | 100 | 1...1000 |
| protocol | pre-set search parameters | 0 |
|
| html | if set, add HTML tags to FASTA outputs | 0 | 0 or 1 |
| seq | query sequence | required, no default | |
| hdr | FASTA header | Query | if used, the URL must be escaped |
| query | query sequence(s) input from textbox | alternative to seq&hdr | must be in FASTA format |
| file | query sequence(s) input by file upload | alternative to seq&hdr | must be in FASTA format |
Fasta, applet and logo modes exit after processing the first query sequence, raw mode after processing one million queries, and the others after 5,000 or 100,000/H queries.
The following examples use a short dummy sequence as query.
| Description | URL |
|---|---|
| Print SANSparallel query form | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi |
| Search database with defaults | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY&hdr=%3EMCINX|MCINXTMP_001238-PA+predicted+protein+(Fragment) |
| Launch Jalview applet embedded in browser | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=applet&html=1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| Launch Jalview Desktop with profile | http://www.jalview.org/services/launchApp?open=http%3A%2F%2Fekhidna2.biocenter.helsinki.fi%2Fcgi-bin%2Fsans%2Fsans.cgi%3Fmode%3Dgapped_fasta%26protocol%3D0%26db%3Duniprot%26H%3D10%26seq%3DLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY%26hdr%3D%3EQuery |
| Launch Jalview Desktop with unaligned hits | http://www.jalview.org/services/launchApp?open=http%3A%2F%2Fekhidna2.biocenter.helsinki.fi%2Fcgi-bin%2Fsans%2Fsans.cgi%3Fmode%3Dungapped_fasta%26protocol%3D0%26db%3Duniprot%26H%3D10%26seq%3DLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY%26hdr%3D%3EQuery |
| Search Swissprot using slow protocol and output 10 hits | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?db=swiss&H=10&protocol=1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| Search Swissprot using very slow protocol and output 10 hits | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?db=swiss&H=10&protocol=2&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| Search Swissprot for 10 hits using very fast protocol | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?db=swiss&H=10&protocol=-1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| Search Swissprot for 10 hits using very fast protocol and output FASTA sequences to browser | HREF=http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=ungapped_fasta&db=swiss&H=10&protocol=-1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY&html=1 |
| Search Swissprot for 10 hits using very fast protocol and output BLAST-like report | HREF=http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=pairwise&db=swiss&H=10&protocol=-1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| Search Swissprot for 10 hits using very fast protocol and output profile to browser in gapped FASTA format | HREF=http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=gapped_fasta&db=swiss&H=10&protocol=-1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY&html=1 |
| Search Swissprot for 10 hits using very fast protocol and output alignment colorized by Mview | HREF=http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=mview&db=swiss&H=10&protocol=-1&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| Search UniRef50 for 1000 hits using very slow protocol and generate a sequence logo | HREF=http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=logo&db=uniref50&H=1000&protocol=2&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY |
| SANSparallel's raw output format. View source to see all. | http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi?mode=raw&seq=LHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNY&html=1 |
Exercises with curl and Perl
These exercises are done in a Linux environment using curl to fetch data from the SANSparallel web server and using local Perl scripts to process the data. The syntax of curl iscurl --form key1=value1 .. keyN=valueN URLThe URL of SANSparallel is http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi. The key/value pairs were described in the previous section.
- Fetch ten FASTA sequences from UniRef50 for a query sequence and pipe them to an alignment program.
- Parse information from SANS raw output. Let's use predicted protein sequences from newly sequenced pathogen as an example.
- Copy the FASTA file dickeya_solani.fasta to your file system.
- Do database searches with SANSparallel server
- Use the Perl script called sansparser.pl to produce tabular output. It will output the columns given in the argument list.The output can be viewed in Excel, for example.
perl sansparser.pl qpid spid qcov scov bitscore desc < sans.raw > sans.tab
- Demonstration of Best Informative Hit approach to protein annotation. We use the database search results in sans.raw from the previous exercise.
- Pipe parsed SANS data to a Perl script called BIH.pl to pick the best informative hit for each query sequence.
perl sansparser.pl qcov scov species qpid evalue desc seq < sans.raw | grep -v 'Dickeya solani D s0432-1' | gawk ' $1 > 0.7 && $2 > 0.7' | cut -f 4- | perl BIH.pl > dickeya_solani.html
- What does the gawk step do, and why do you think it is inserted into the pipeline?
- Open the resulting HTML table dickeya_solani.html in a web browser. Details for each protein can be checked by clicking on the link to a database search by SANSparallel.
curl --form mode=ungapped_fasta --form db=uniref50 --form H=10 --form seq=MLFHTDDLQPIFETENRPDNFSERVSETQSDTPKGGETQPEIVTVGKEKKKGGKTFPCAQCGREFAHKNSLAYHTLMHGDKQQACRDDYRPCKCDECGRQFRQWSDLKYHKASLHSDKKQFKCDFCGKEFARRYSLSVHRRIHTGEKNYKCEYCNKAFRASSYRLIHMRTHTGSKPYKCPQCDKGFRVSYDLQRHMHIHEKVRVKADDQKKTKDTKEKKQTITKTNEEKKEPESPSSEQIKSENRLPMLKSLLDKKPAKQSKKSPKKAPNVTVQNKIDEQFDEEIFDTRQDPYKFKEVYTNEKEFSNISHKFDRTDERELENLRSIKIPQIGETEDRNYSRENTDGKMQVFTQIDKGKEYSGPIVTNVVSLSDIRNLEREVLREPRVEIQGDGLENGFFERLSAFYNISAI http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi | /usr/local/bin/muscle3.8.31_i86linux64 -html -out muscle.htmlView the result muscle.html in a web browser.
curl --form mode=raw --form file=@dickeya_solani.fasta http://ekhidna2.biocenter.helsinki.fi/cgi-bin/sans/sans.cgi > sans.rawIf the raw data file ends with Elapsed time, then you know that the data was transferred completely.
Troubleshooting
| Problem | Solution |
|---|---|
| ERROR in connect | Client failed to connect to SANS server. The server is down or only starting up. Try again later. If the error persists longer than a day, contact SANS team. |
| Java applet doesn't work. Java warns about expired certificate. | Check your security settings as in Exercise 1.2 |
| Browser becomes unresponsive. | It's a problem with your browser's memory. Close the browser window and restart. Input fewer sequences, or request fewer hits in output. |
| Loading the page took a long time to finish and in the end I only got a white page. | If server load is very high, it is possible that the timeout for keeping socket connections alive expires (~30 seconds). |
| How can I download results for my transcriptome data set containing one million predicted proteins? | Use programmatic access to bypass browser. |
| Result says 'Illformed query'. Result says 'Unknown output mode'. | Check the syntax of external links, or use the query form. |
| Bitscores and e-values in BLAST-like report don't match the values in Table view. | The two outputs are generated by different programs. Table view results come from SANSparallel. BLAST-like report comes from fasta36 against a small database consisting of the hits found by SANSparallel. |
| Table view has more hits than BLAST-like, gapped FASTA or Mview outputs. | That's because the latter go through pairwise alignment by fasta36, which computes e-values differently and only passes through those hits that get an e-value below one. |
| Mview takes a long time | Yes it does, doesn't it. |

